DArTView¶
About¶
DArTView is a KDXplore plug-in for marker data curation via metadata filtering. Its primary goal is to overcome tedious manual calculation of marker data through common spreadsheet applications, steps such as:
-
Creating formula for a statistic (e.g. Marker Call Rate)
-
Extending formula to entire dataset
-
Sorting data via calculated statistic
-
Visualising calcualted staistic (e.g. via scatter plot)
-
Subsetting dataset based on determined threshold
-
Repeating steps 1-4 to examine effect on other statistical distributions
Supported Formats¶
DArTView supports multiple nucleotide genotypic data types of DArT and Hapmap marker data formats:
-
DArT
DArT Counts
DArT SNP Two Row
DArT Silico SNP
DArT Silico Counts
DArT SNP One Row
-
Hapmap
Two Letter
SSR
Codominant
Dominant
In this user guide, these formats will be refered to as ‘marker data’
Contact¶
If you experience any issues or have feedback, please contact kdxplore@diversityarrays.com
User Guide¶
This user guide provides an overview of DArTView, as well as information on importing files and graphing and filtering data.
To begin, see the below section on importing marker data files.
Downloading and Launching the Application¶
Prior to using the application, KDXplore requires Java to be installed on your computer. We recommend installing from https://www.java.com/en/download/.
If you have Java installed, you can download KDXplore-DArTView from http://software.kddart.com/kdxplore/dartview/.
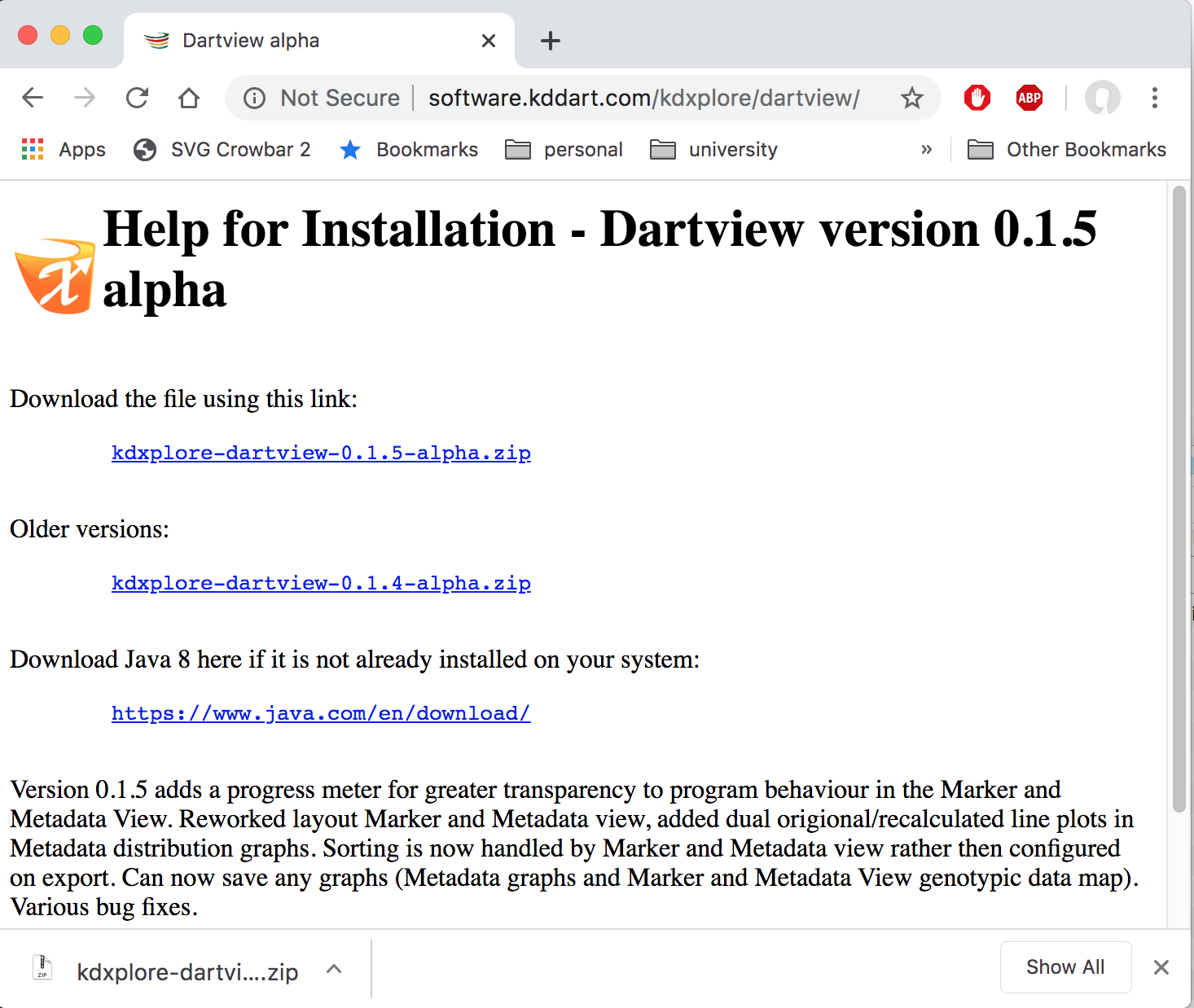
The application is bundled as a ZIP archive which will need to be extracted after downloading:
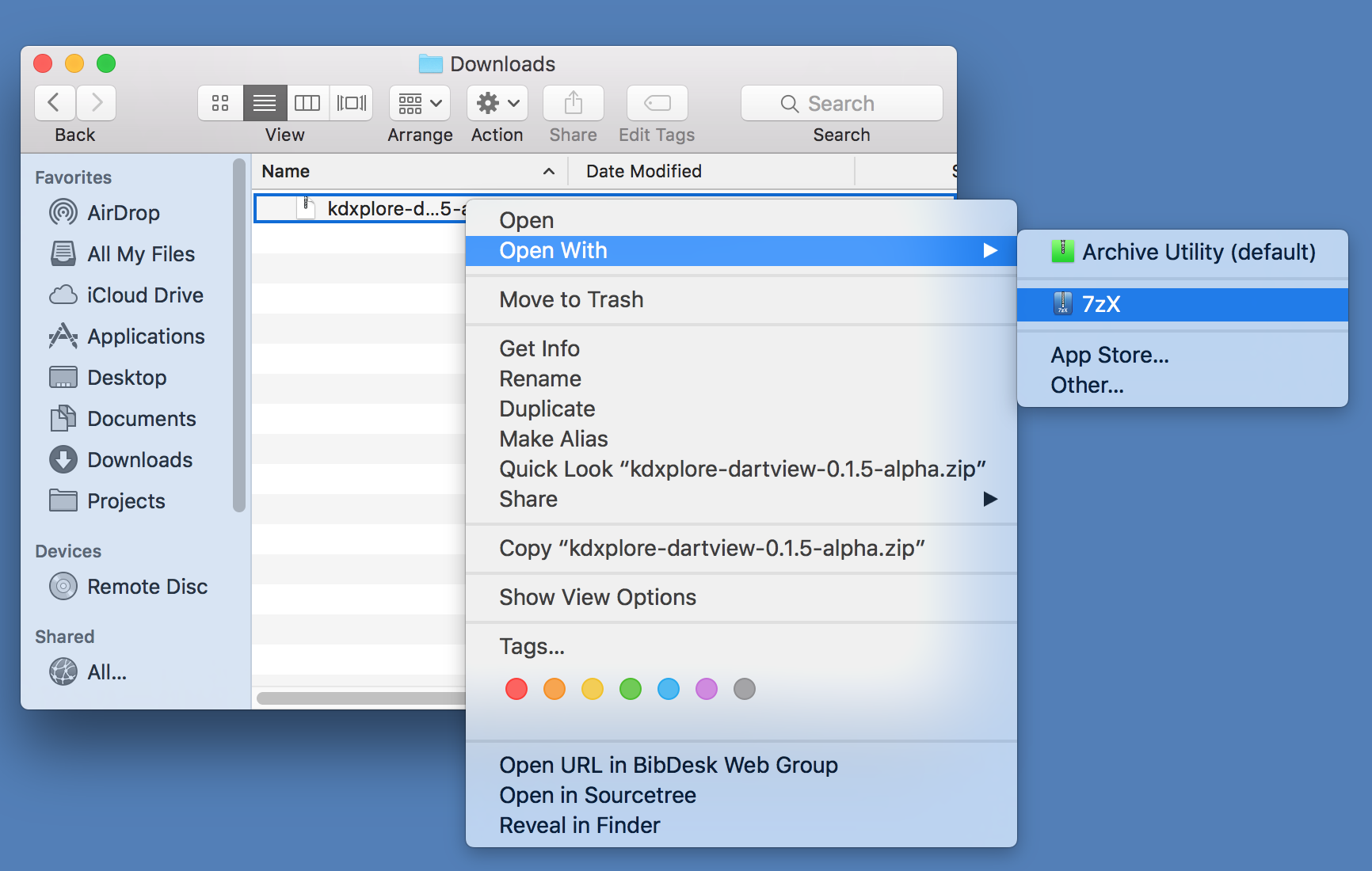
To launch, open kdxplore.jar with Java:
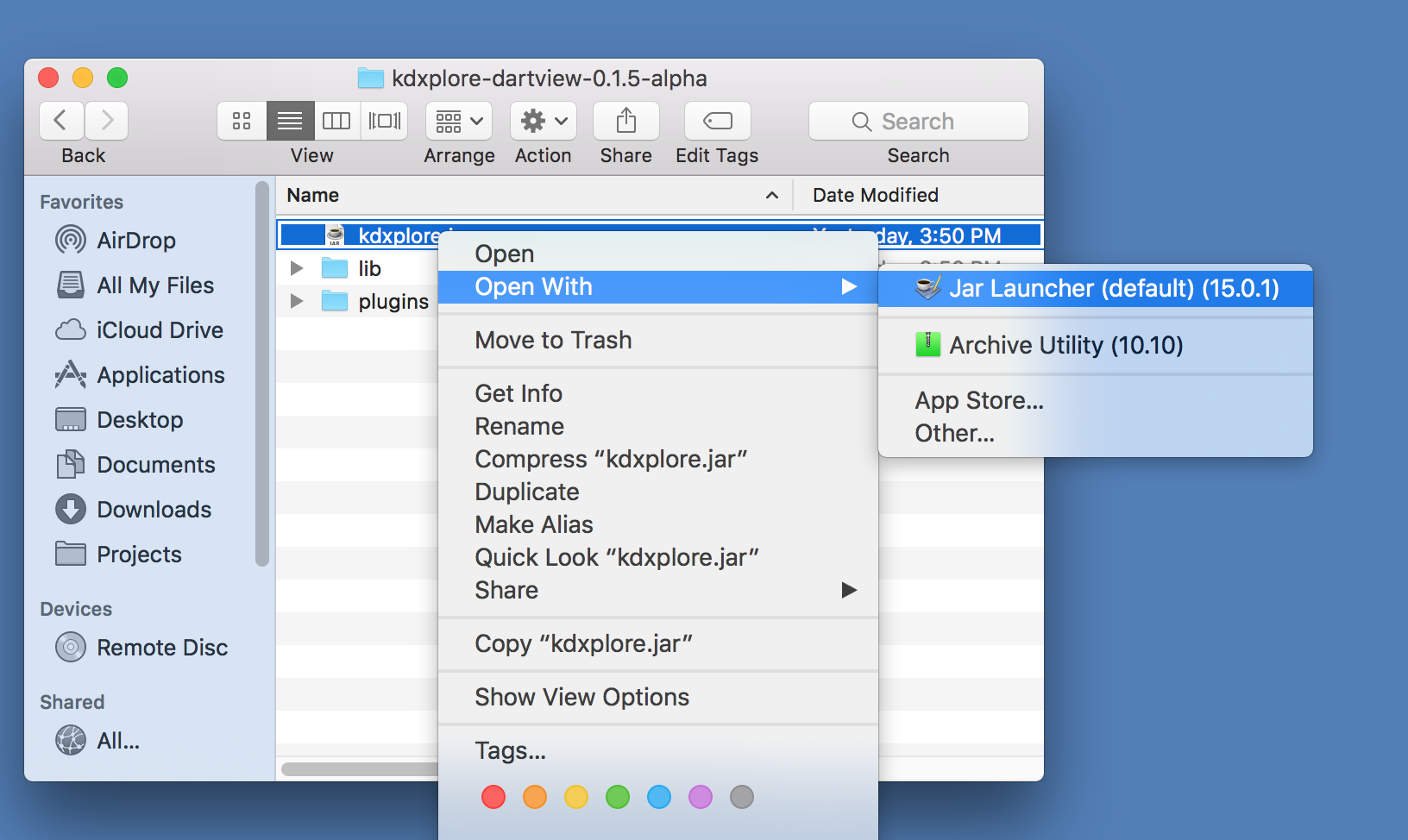
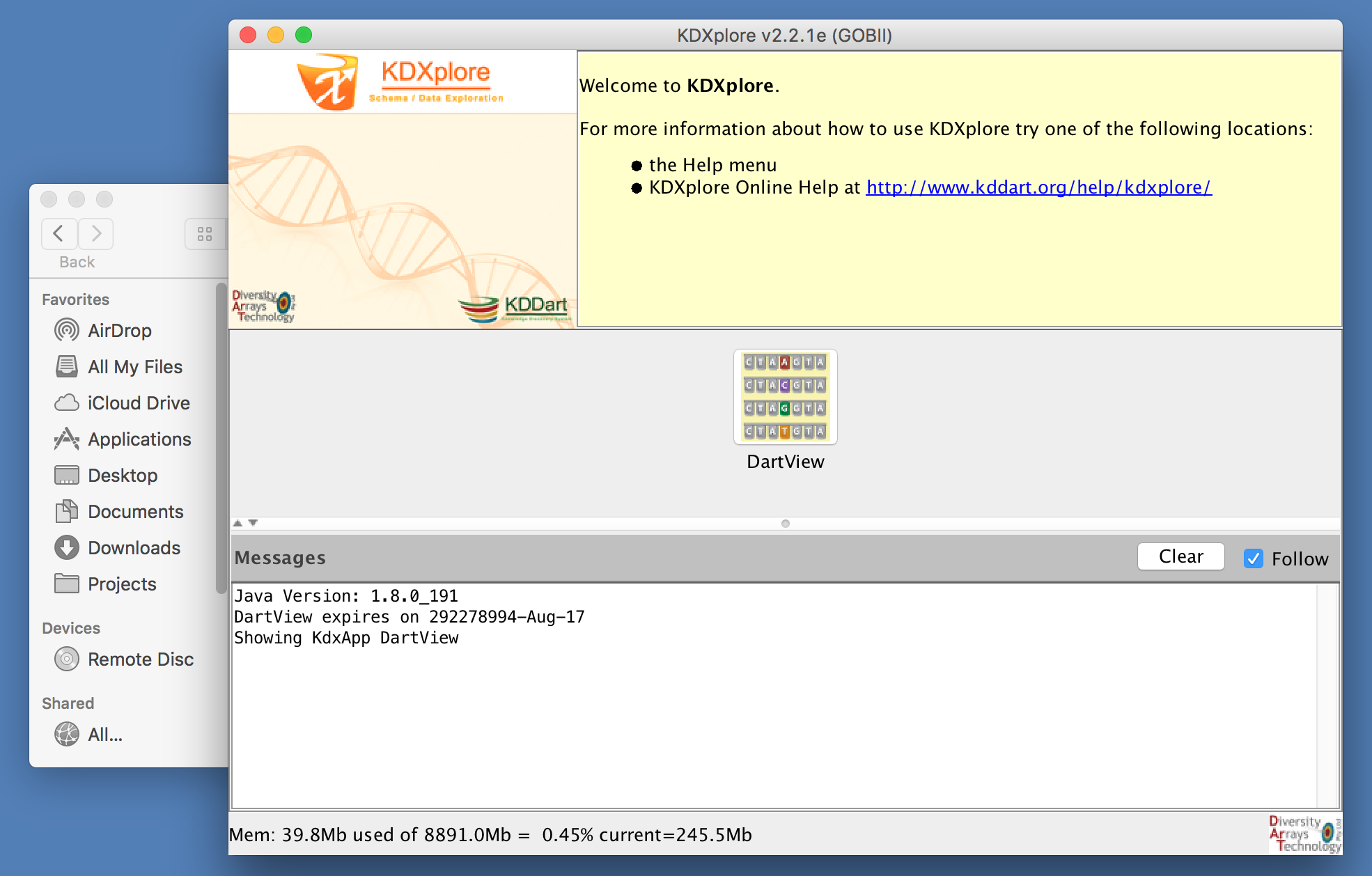
This should load the KDXplore Main Window which houses all available KDX plugins. For this KDXplore version, we just have the DArTView plugin.
To launch DArTView from the KDXplore Main Window, just click the DArTView icon.
This should open DArTView, ready to import a new or recent marker data file:
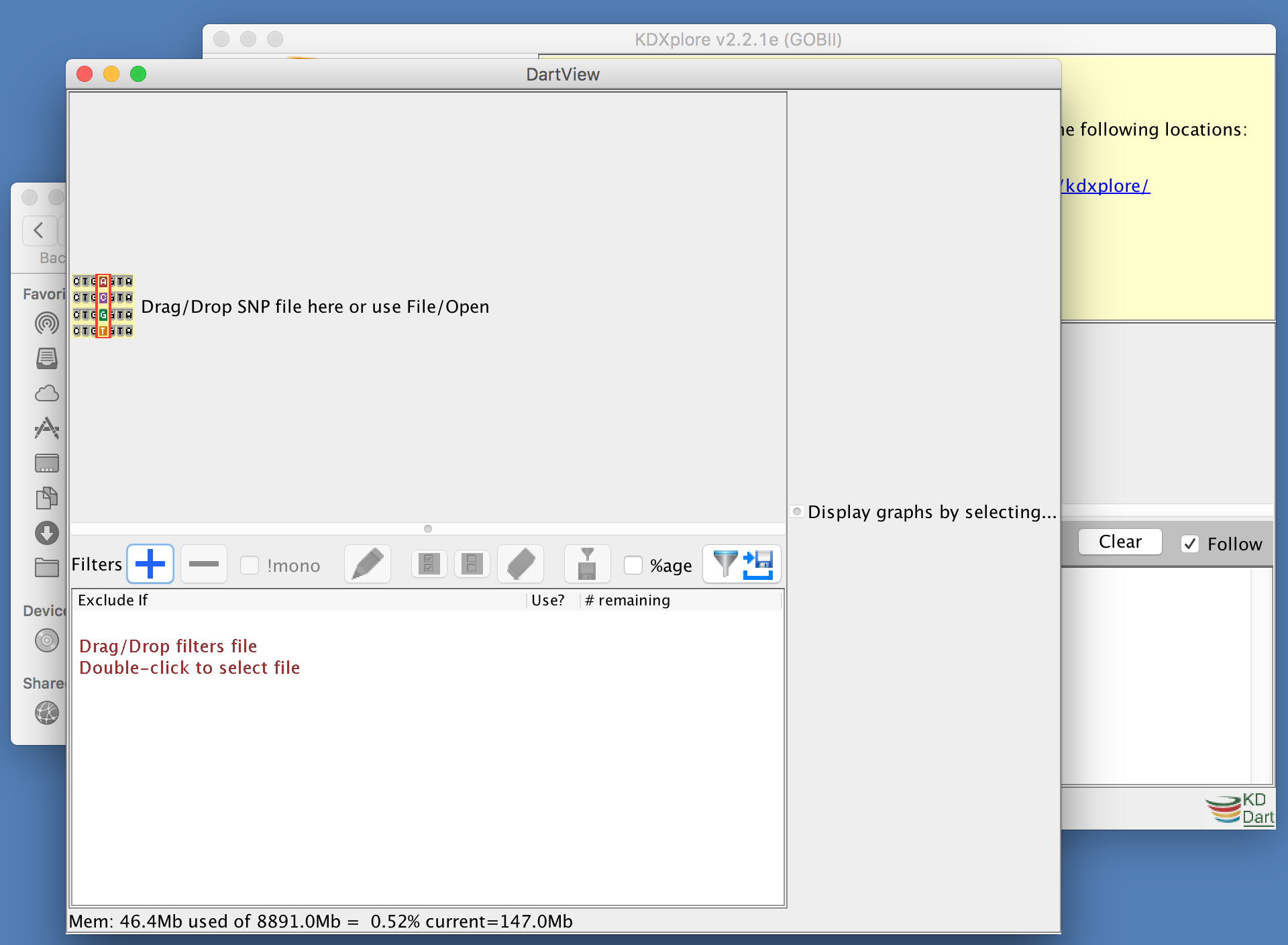
Importing a Marker Data File¶
The first step to working with any data in DArTView is to import a marker data file. This section demonstrates how to import marker data files into DArTView.
To import a marker data file, you can either:
-
Open DArTView and locate the marker data file that you would like to import. Click+drag the file to the File Panel in DArTView.
-
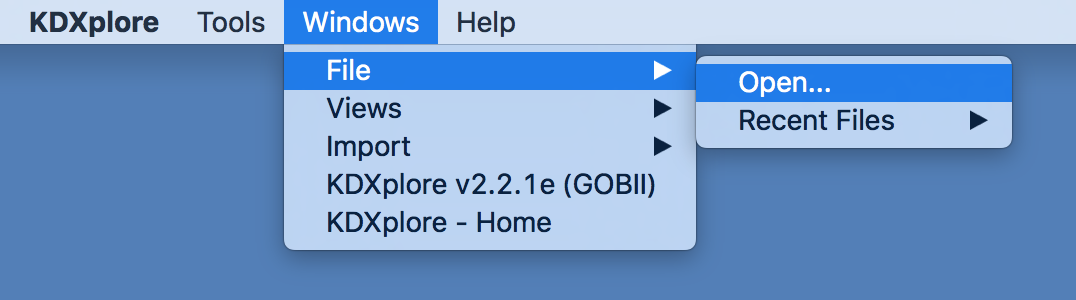
Alternatively, open dartview via the application menu: * Windows->File->Open…
DArTView will attempt to automatically recognize your file and proceed to open it, but in case of ambiguity you’ll be prompt with the Import Marker Data Wizard. The wizard will require users to specify their marker data’s format, marker type, Report format (DArT only) and to identify sample metadata.
The next section details the steps of the Marker Data Import Wizard:
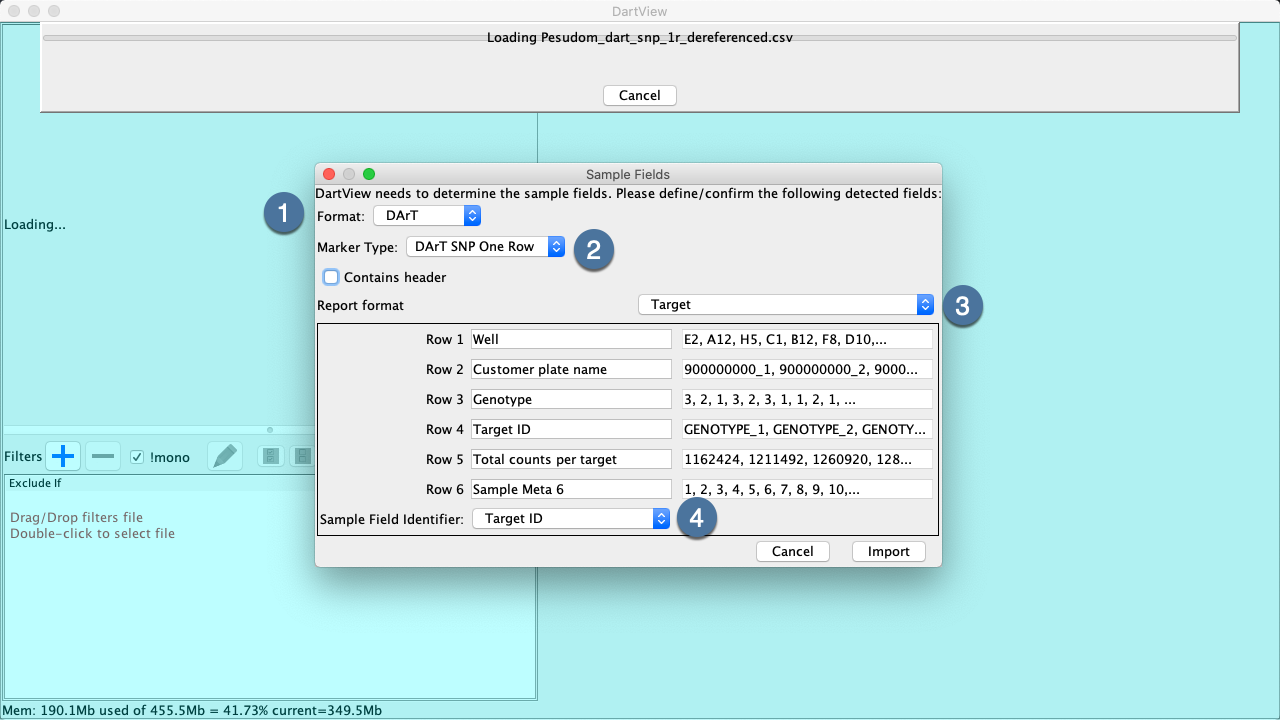
Note: Incorrect configuration can lead to incorrect metadata calculation
DArTView Main Window¶
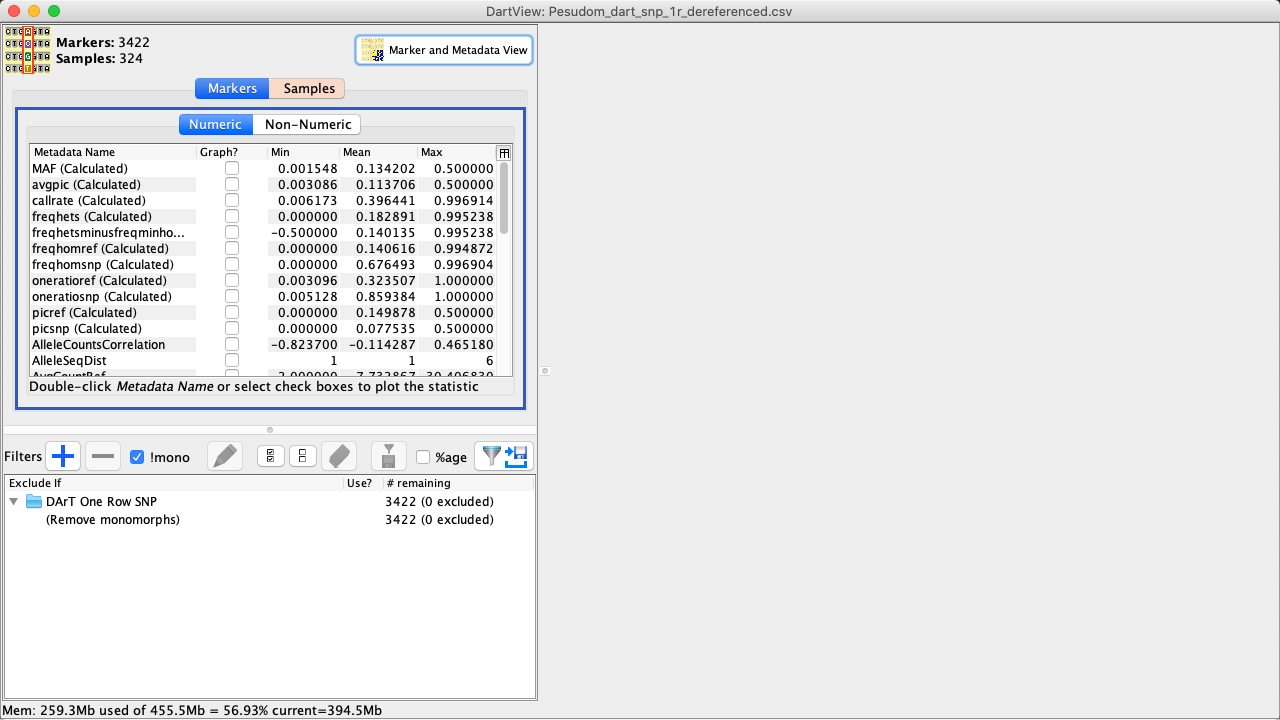
The DArTView Main Window displays an opened file, as well as filters and graphs that have been applied.
|
Item |
UI Element |
Description |
|---|---|---|
|
Metadata Panel |
Lists metadata that is organised into markers/samples with subsections numeric/non-numeric each. Checked metadata are displayed as a graphs in the Graphs Panel. |
|
|
Filters Panel |
Lists all active/unactive filters that have been added to the file and provides options for managing these filters. |
|
|
Graphs Panel |
Displays scatter and histograms plots based on the filters applied to a file. |
Viewing Metadata Statistics¶
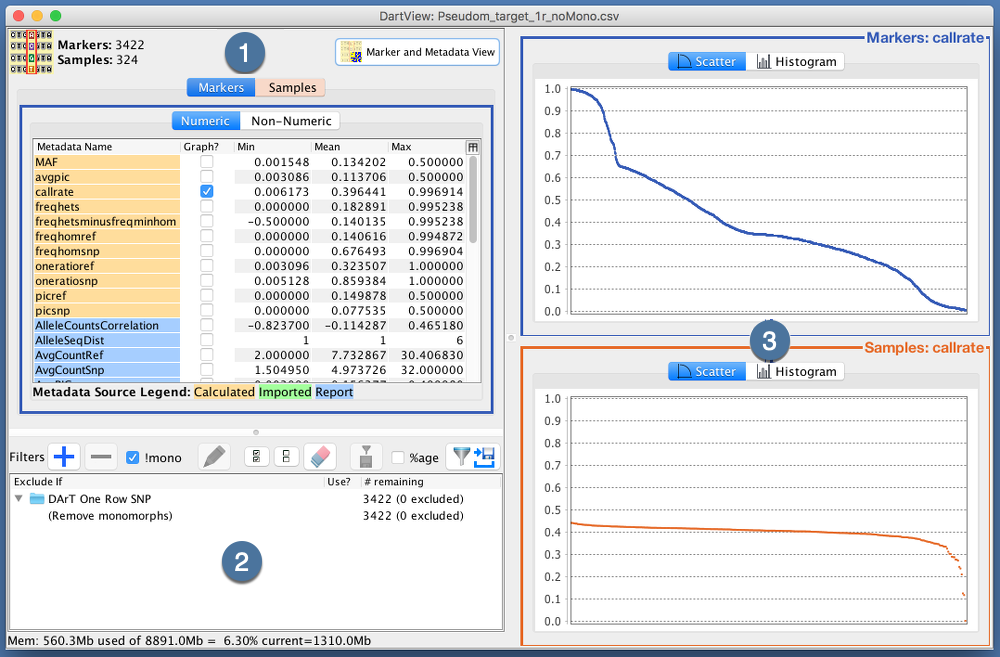
Data can be viewed after it is loaded from a marker data file. The options for viewing data are outlined in the image and table below:
|
Item |
UI Element |
Description |
|---|---|---|
|
Report Counts |
The total number of markers and samples are displayed here. |
|
|
Metadata Tabs |
Select either the Markers Tab or the Samples Tab to view metadata on markers or samples from the report. The Numeric Tab and Non-Numeric Tab allows the user to view numeric and non-numeric metadata. |
|
|
Stats Button |
The Metadata Panel in the above image displays only basic metadata stats. Select the Stats Button to choose the all stats option which displays all metadata stats available. |
|
|
Marker and Metadata View Button |
This button will open a window called the Marker and Metadata View where data can be graphed. See the section below for more information. |
|
|
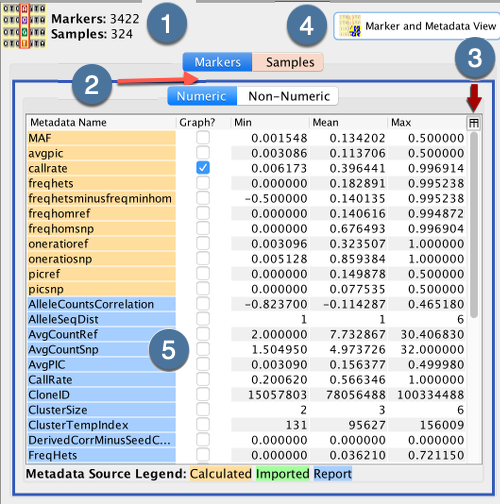
Metadata Panel |
This panel displays metadata according to what tabs the user has selected to view. Select the checkbox or double-click on any of the rows to display further information on the metadata. This is demonstrated in the image below where the callrate is selected and displayed. |
|
Statistic |
Description |
|---|---|
|
Min |
The minimum value for this metadata statistic |
|
Mean |
The mean value for this metadata statistic |
|
Max |
The maximum value for this metadata statistic |
|
Extended: Q1 |
First interquartile range. The “middle” value in the first half of the rank-order data set |
|
Extended: Median |
Second interquartile range. The “middle” value of the rank-order data set |
|
Extended: Q3 |
Third interquartile range. The “middle” value in the second half of the rank-order data set |
|
Extended: Variance |
The sample variance of the statistic: :raw-latex:`\frac{1}{N-1}\sum_i^N (\overline{X} - X_i)^2` |
|
Extended: Std Dev |
The sample standard deviation of the statistic: :raw-latex:`\sqrt{\frac{1}{N-1}\sum_i^N (\overline{X} - X_i)^2}` |
|
Extended: Std Err |
The sample standard deviation of the statistic: :raw-latex:`\frac{\sqrt{\frac{1}{N-1}\sum_i^N (\overline{X} - X_i)^2}}{\sqrt{N}}` |
|
Statistic |
Description |
|
Values |
Displays some entries of the metadata |
|
Source |
Description |
|---|---|
|
Calculated |
Metadata has been calculated using DArTView’s internal set of calculators for this data format |
|
Report |
Metadata was imported from report |
|
Imported |
Metadata was imported via “Import Metadata Wizard” |
Viewing Graphed Metadata¶
Each metadata row can be viewed in more detail in different graphs located in the Graphs Panel. Select the checkbox or double-click on any of the rows to display more information in the form of scatter plots and histograms of the data.
The images below are examples of the callrate being graphed as both a scatter plot and a histogram. The data is displayed in descending order as that is what order it was designated in the Metadata Panel. There are also checkboxes along the bottom on the scatter plot which provide options for marking summary statistics of the data. The median is marked in this example and can be seen with the blue line through the scatter plot.
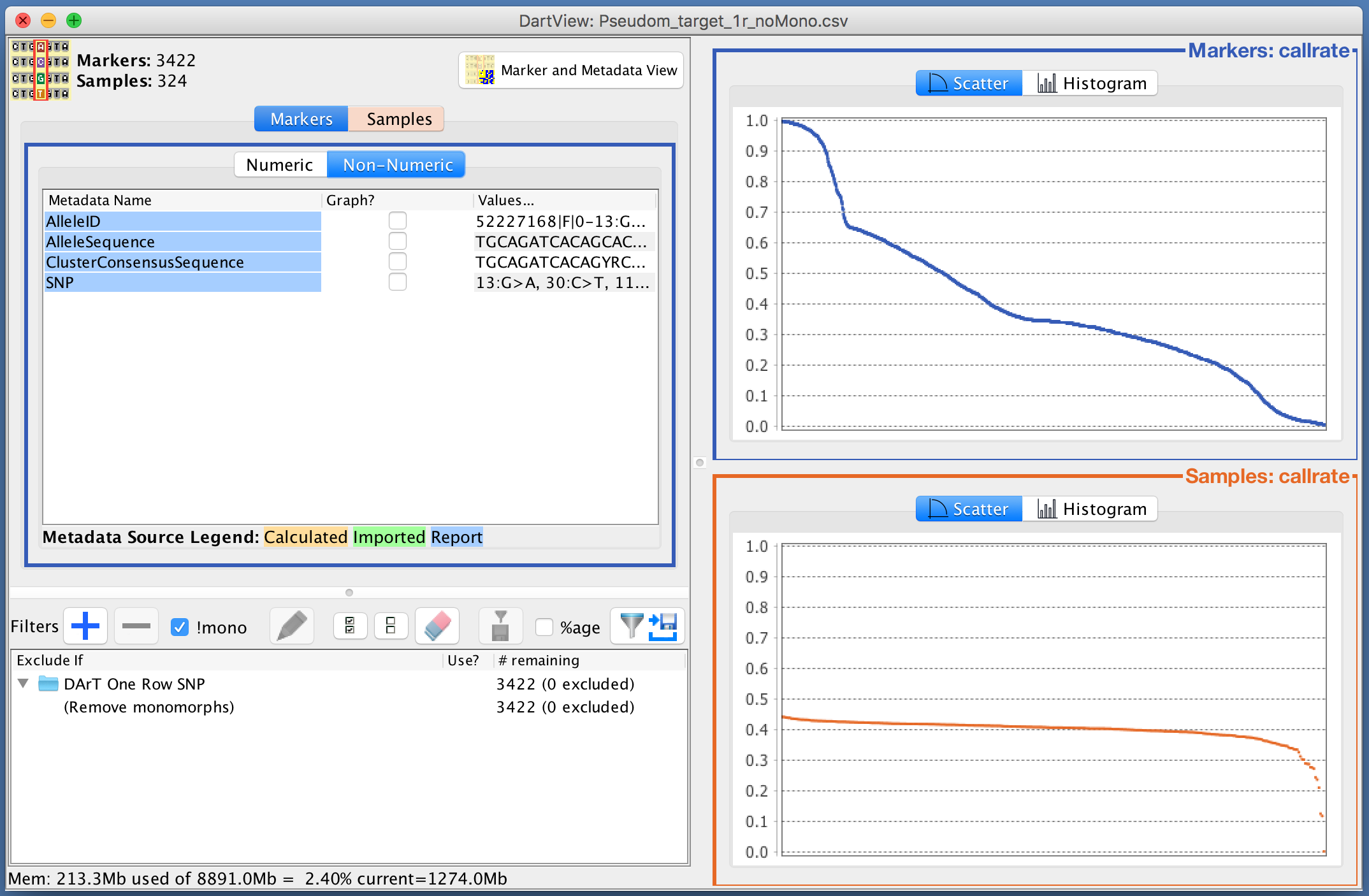
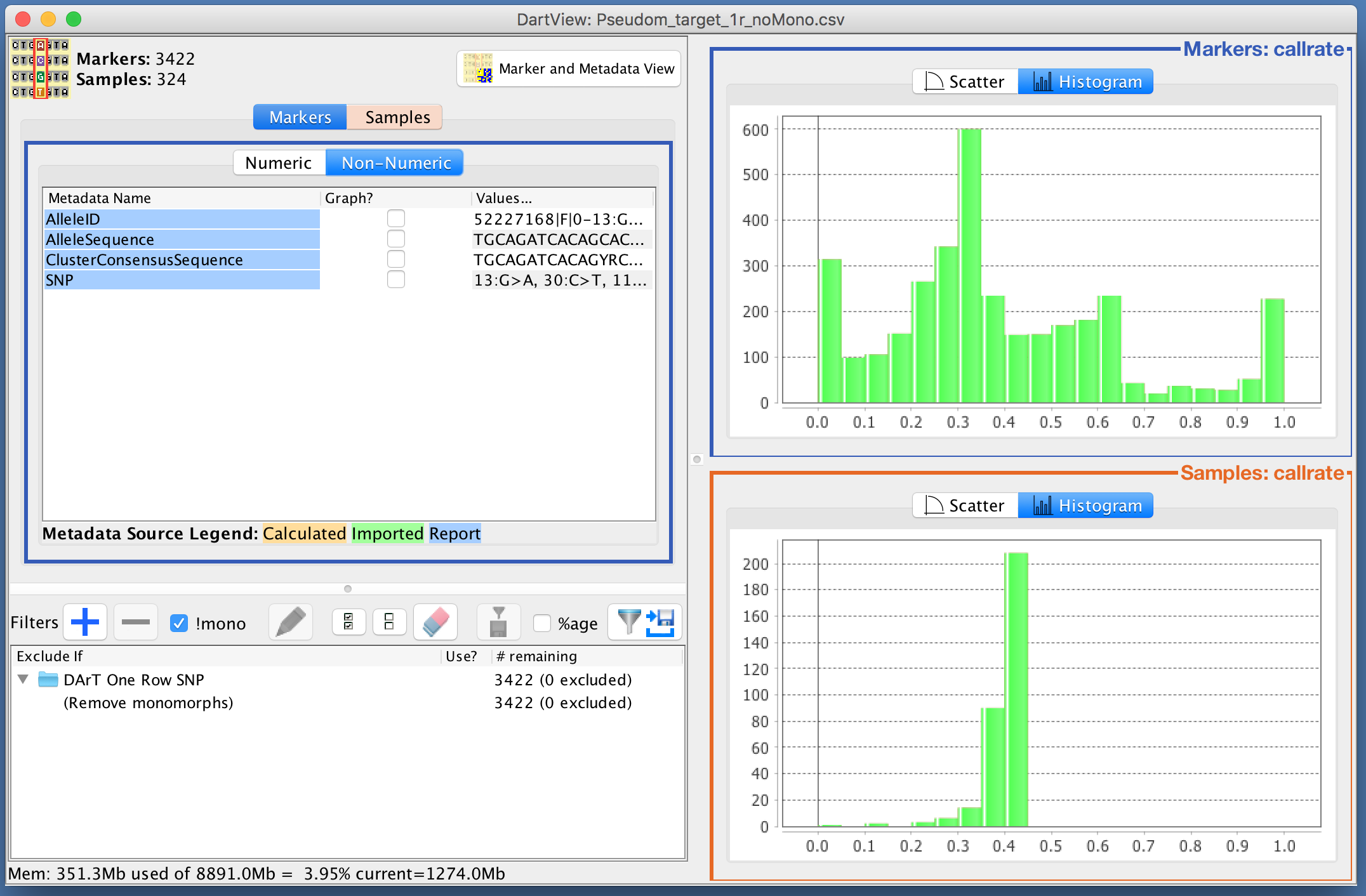
Adding Filters¶
Filters can be used to refine the data set that is viewed in DArTView and can be saved for reuse. Filters can be either created or loaded from a previously saved filter file.
Filter Panel¶
The Filter Panel displays all active and unactive (Checked/Unchecked) filters and provides options for managing those filters. The filter panel will switch with the Markers or Samples tab. The image and table below outline the options that are available:
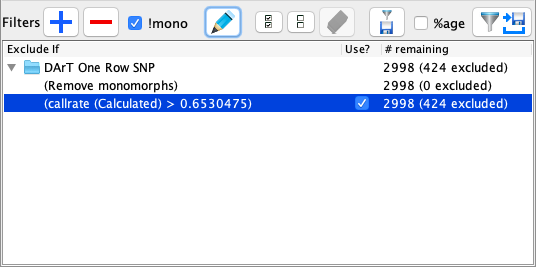
|
UI Element |
Description |
|---|---|
|
Displays the Add New Filter Window which provides options for creating a new filter. |
|
|
Removes the selected filter. |
|
|
Remove Mono Checkbox (Markers only) |
This is a special filter which is applied only after standard filters (Both markers and samples) are applied. A monomorphic marker is defined as a marker with only one allele visible across the samples (Ignore missing values). Such markers are considered uninformative and are often redundant in downstream analysis. The Remove Monomorphs Filter is always available by default and can’t be removed. However, this checkbox allows the user to enable/disable this filter. |
|
Opens the Edit Filter Window for editing a selected filter. |
|
|
Enables all filters in the Filters Panel. |
|
|
Disables all filters in the Filters Panel. |
|
|
Disables all filters that are not applicable to the marker data file that is currently open. |
|
|
Export Filters Button |
Provides options for exporting selected filters to a separate file so that they can be reused again. |
|
Percentage Checkbox |
By default, each filter will display the number of data points that are excluded if the filter is active. This checkbox will display the number as a percentage rather than a count. |
|
Export Data with Filters Button |
Provides options for exporting data with the filters that have been applied. |
Exporting and Importing Filters¶
Exporting¶
Saving a filter will create a file with saved filters from the current session. This can used for future use.
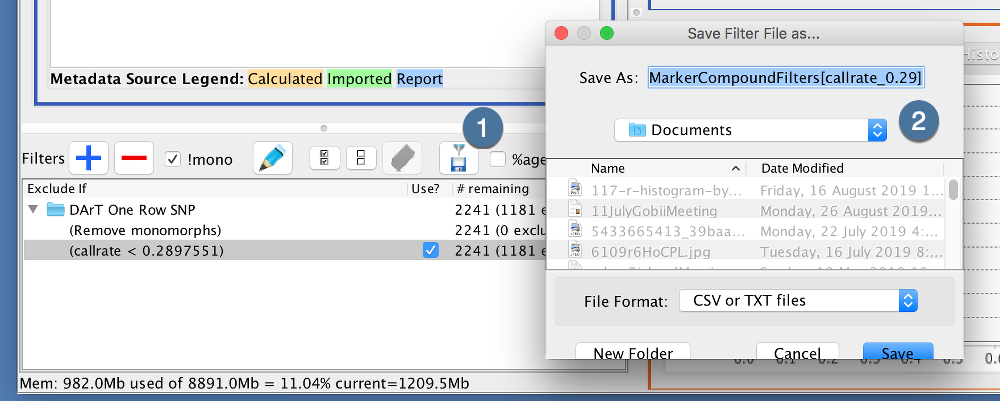
|
1. |
After creating your filters, click the Export Filters Button |
|---|---|
|
2. |
Select a location to save your filter file |
Importing¶
Importing a filter file is performed by dragging the filter file from file explorer and dropping into the Filter panel
Creating New Filters¶
There are two approaches to creating new filters. The first is via the Filter panel and the secondary via the graph.
Creating a Filter in the Filter Panel¶
Filters can be created in the Filters Panel without needing to view any graphed data. See the instructions below for more information:
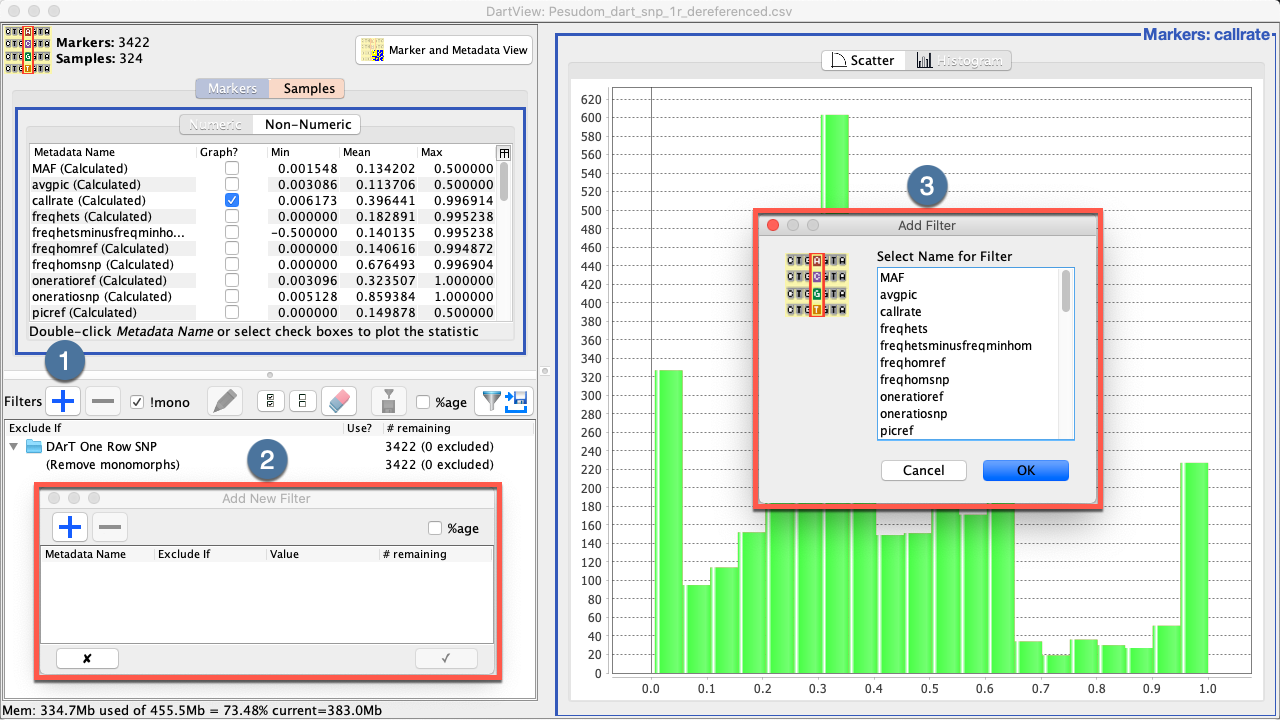
|
Step |
Action |
|---|---|
|
1. |
Select the Add Filter Button |
|
2. |
Select the Add Filter Button |
|
3. |
Choose the metadata for the filter then select the Ok Button to finalise the selection. The Add New Filter Window will be updated as seem in the image below. |
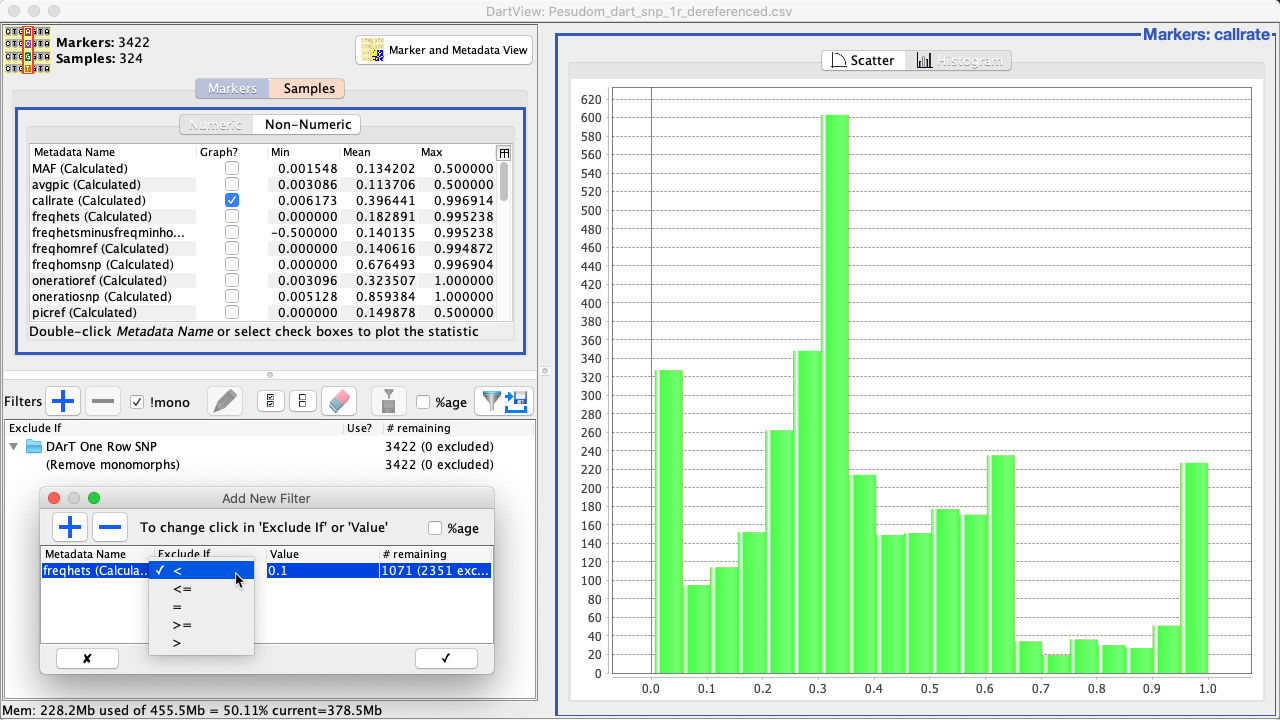
|
4. |
The filter condition can be edited by double-clicking the Metadata Name, Exclude If (such as in the above image), and Value fields. |
|---|---|
|
5. |
Select the Confirm Button (the tick) to finalise the creation of the filter. The graph will be updated such as in the image below. |
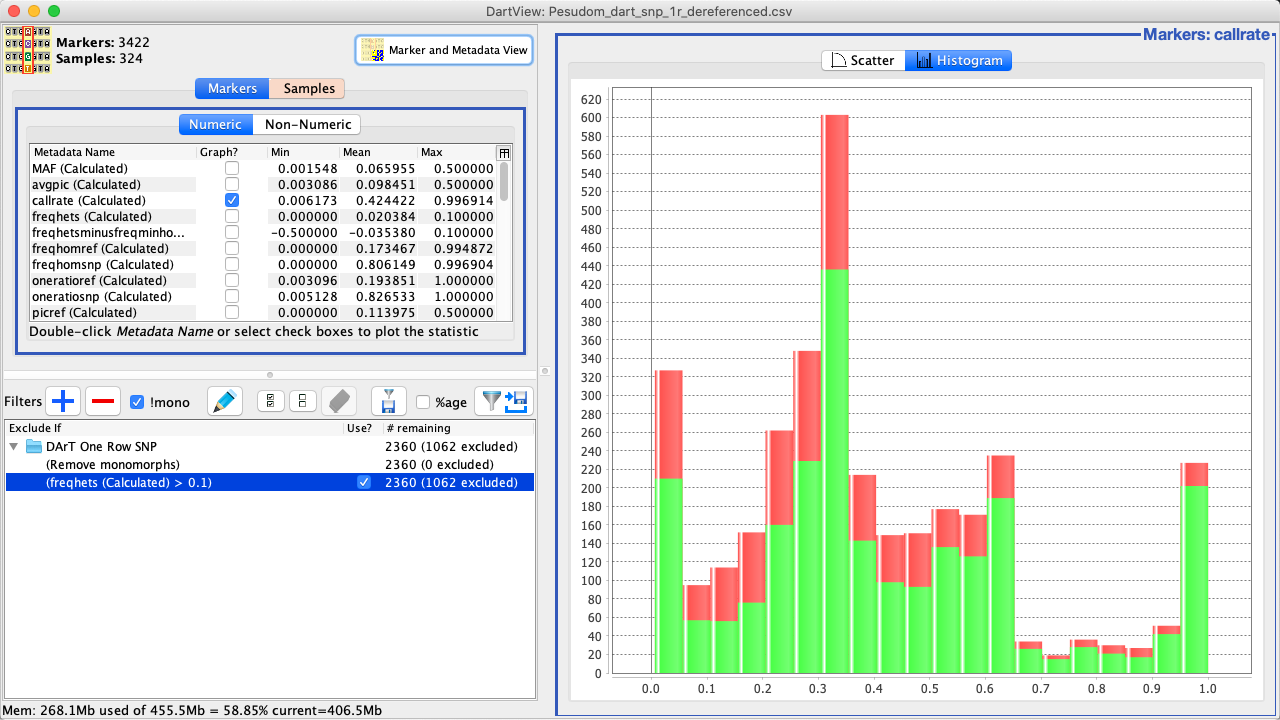
Tip
Both the scatter plot and histogram will be affected by applied filters.
Creating a Filter in a Histogram or Scatter Plot¶
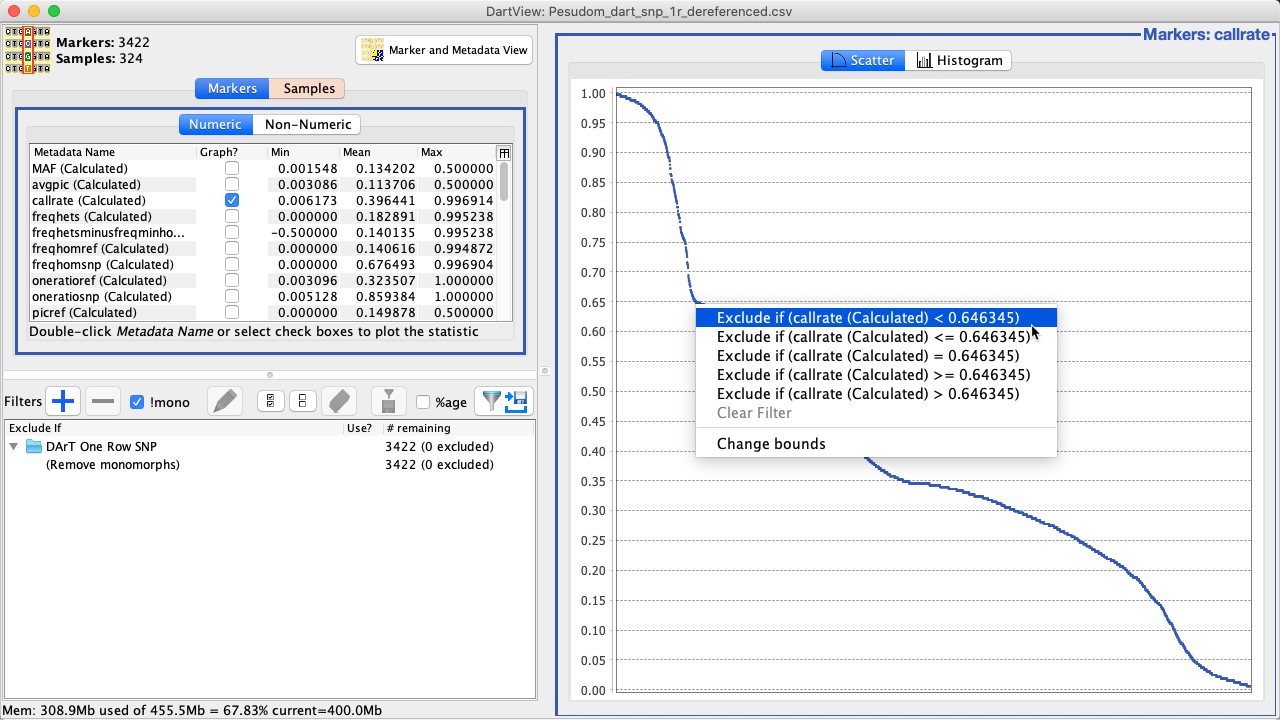
New filters can also be created within a histogram or scatter plot. This allows users to filter based on the graph they are viewing. See the instructions below for more information:
|
Step |
Action |
|---|---|
|
1. |
Open a histogram or scatterplot from the Metadata Panel by selecting the relevant checkbox. |
|
2. |
Right+click in a section within the graph that you want to place a filter (such as in the above image). Filtering (and other) options will be presented. |
|
3. |
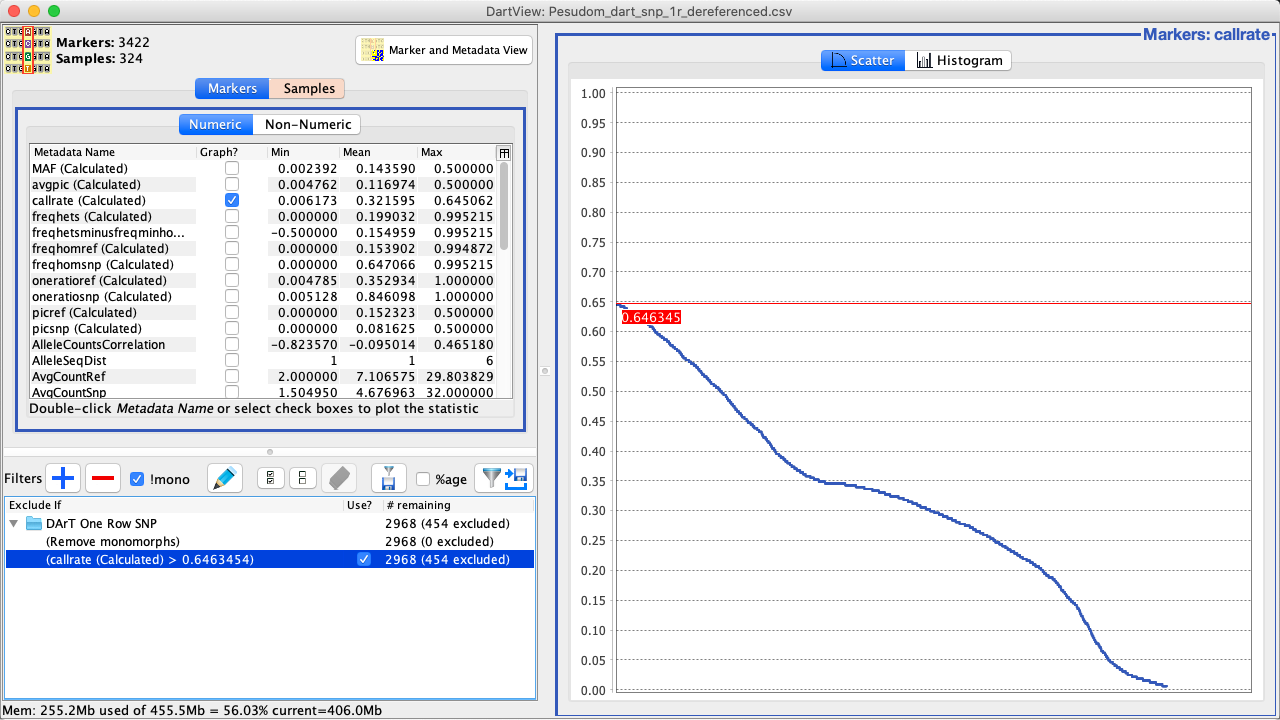
Select a filtering option. This will add and apply the filter immediately as seen in the image below. |
Importing Additional Data Files¶
It is also possible to import additional marker or sample metadata files which are appended to an existing, open marker data file. See the instructions below for how to import additional data:
Additional metadata can be imported in the form of a CSV table data. This can be with or without a header.
To begin, choose to import either Marker metadata or Sample metadata via the menu:
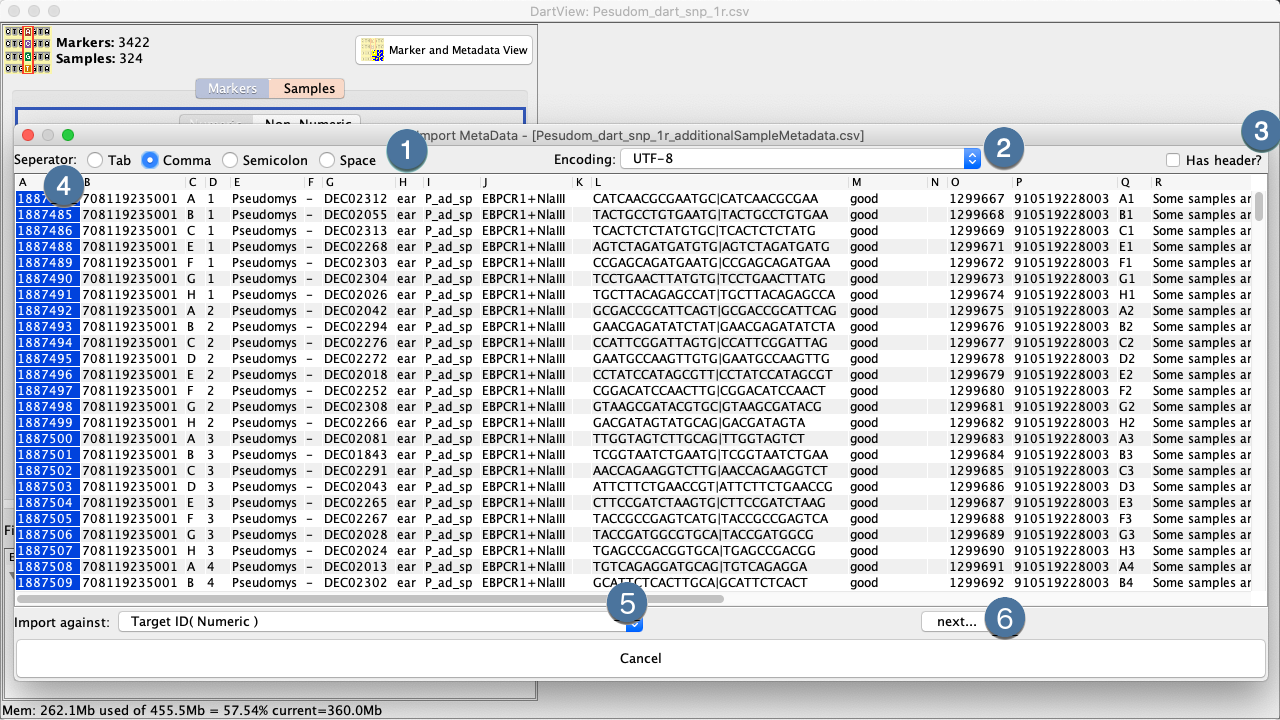
This will prompt you to specify the CSV to import.
|
1. |
Select the delimiter of your CSV |
|
2. |
Specify the encoding. Typically, CSVs are UTF-8 |
|
3. |
Specify whether the file contains a header or or not. Headerless columns with be identified by column ID. |
|
4. |
Click the header of the field that will be used to align against an existing metadata field. This field must have unique values to ensure the imported metadata can be mapped (Note: Your metadata file does not need to be in the same order as your Marker Data File). |
|
5. |
Select which exiting metadata field in DArTView to align your new data against. Similarly, this field must have unique values |
|
6. |
Select the Import Button to finalise the import of the file which will take you to the DArTView Main Window which should now look something like: |
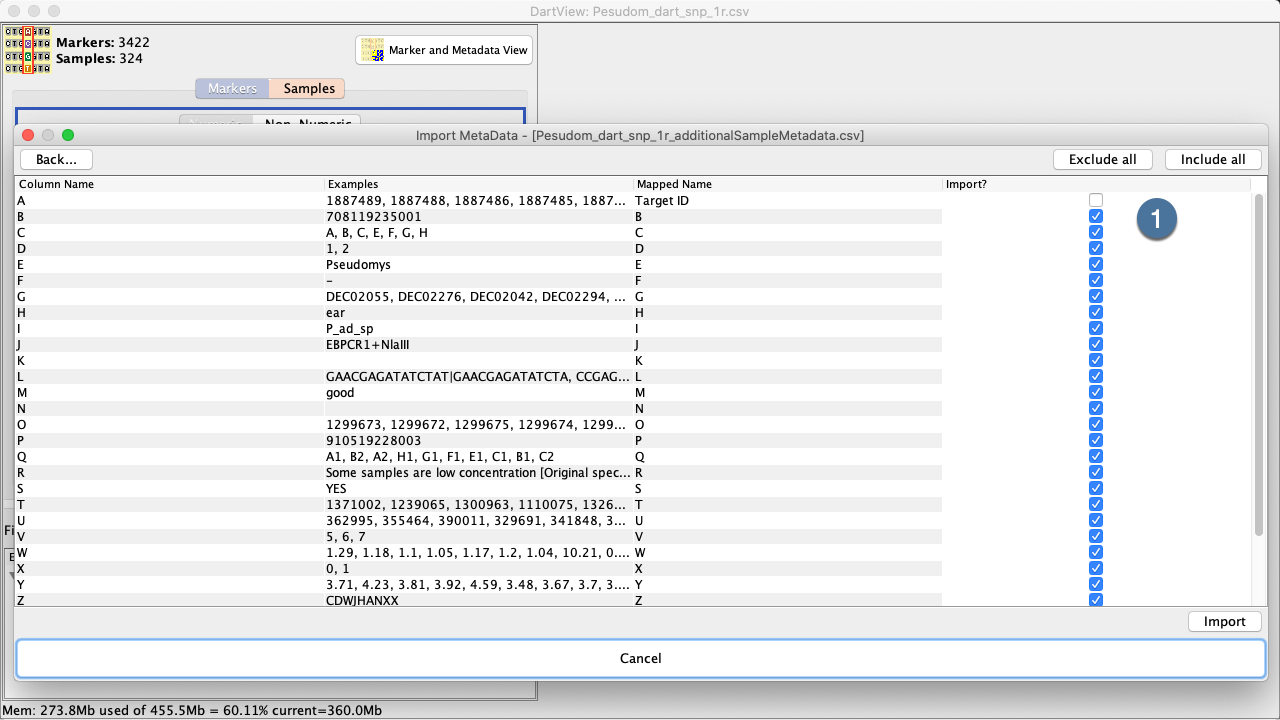
The Marker and Metadata View¶
The Marker and Metadata View provides a full coloured visualisation of the genotypic data. Each genotype data point has a representative color. Colouring is configured by the “Choose Colours” dialog:
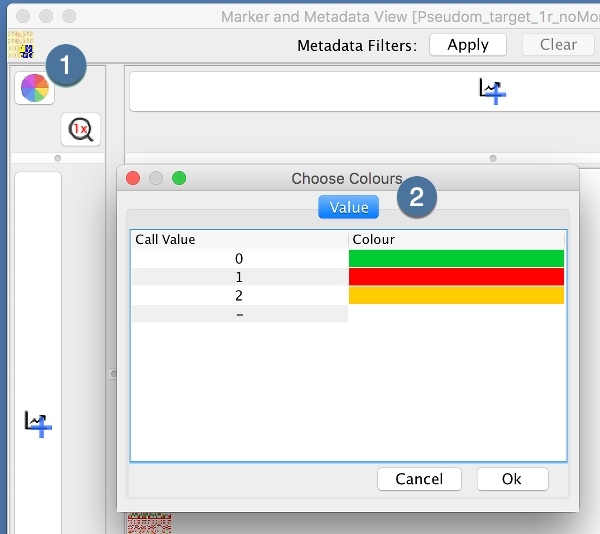
|
Step |
Action |
|---|---|
|
1. |
Open the “Choose Colours” Dialog by clicking the color wheel |
|
2. |
Set each color of each “Call Value” by clicking the “Colour” |
Clicking “Ok” will update the genotype data points displayed.
The Genotypic Data Map¶
The Genotypic Data Map provides a contrasting image of the marker data before and after filtering. Along with sorting capabilities, this feature provides a quick perspective of the dataset.
In the image below, the Genotypic Data Map is sorted by marker call rate and a species sample metadata which had been imported via the “Import Additional Metadata Wizard”.
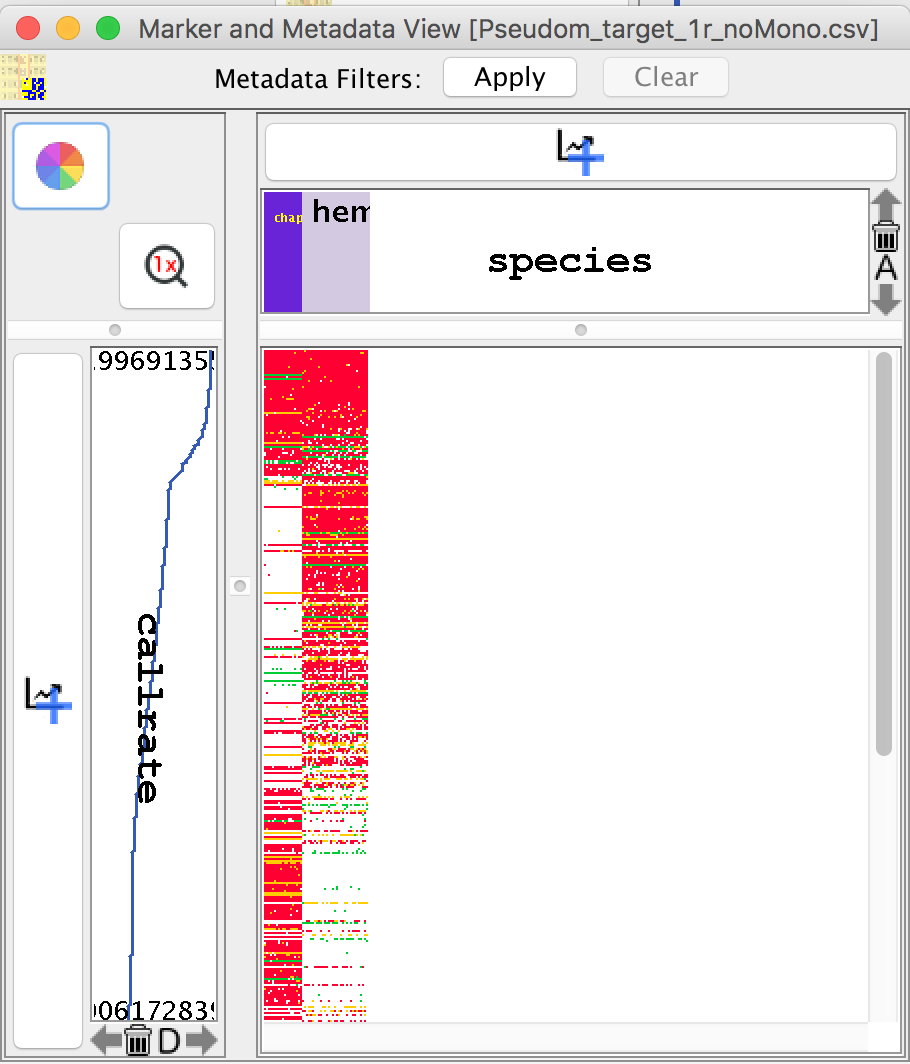
Graphing Data in the Marker and Metadata View¶
Metadata distributions of either Markers or Samples can be added alongside the Genotypic Data Map.
See the below table for information on how to graph data in the Marker and Metadata View.
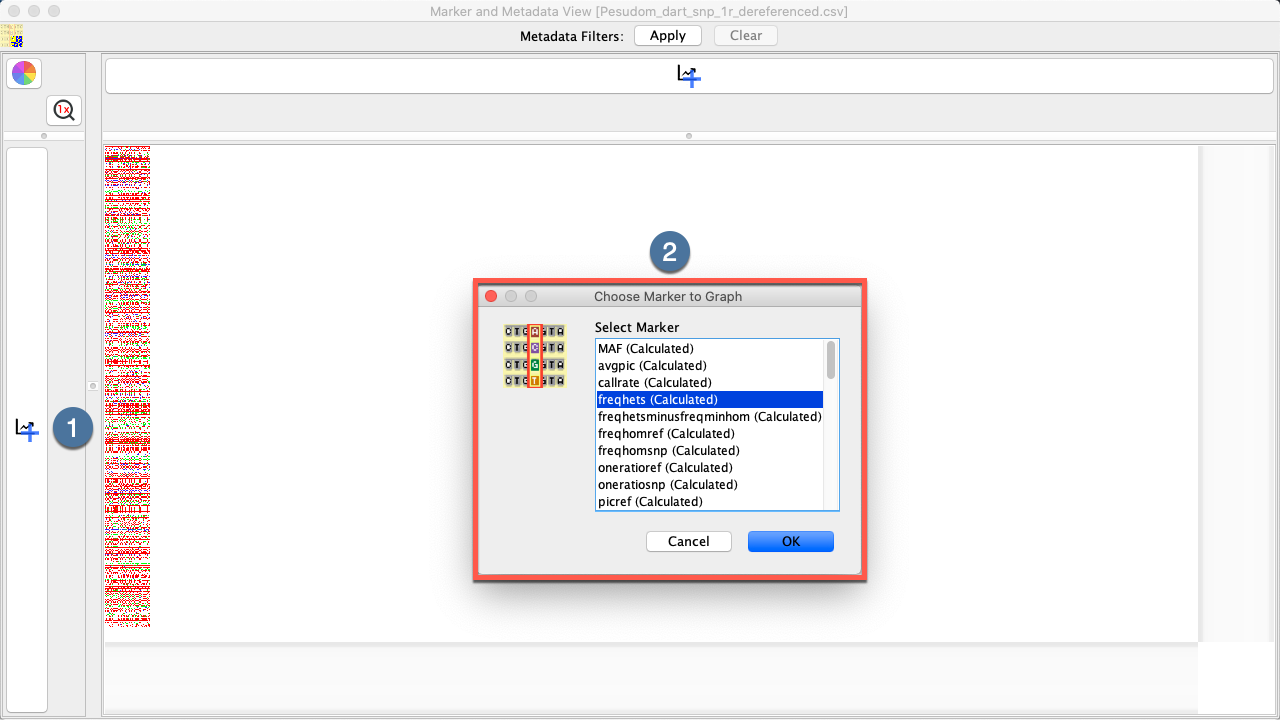
|
Step |
Action |
|---|---|
|
1. |
Open the Marker and Metadata View Window to view genotypic data. |
|
2. |
Select the Add Graph Button |
|
3. |
Select a graph option and then the OK Button. The new graph will be created and applied to the appropriate location. The below image shows that the example graph looks like. |
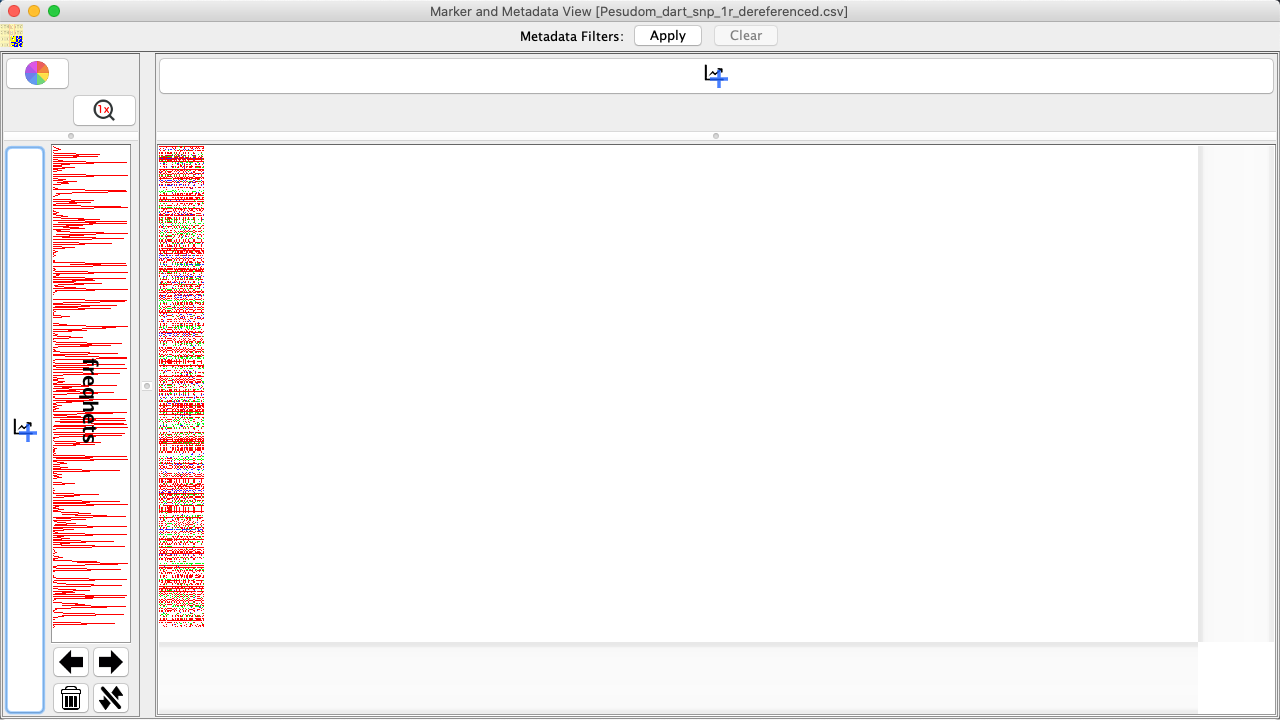
Graph and Viewing Options¶
There are display options available once a graph has been added. The following options are available:
|
UI Element |
Description |
|---|---|
|
As multiple graphs can be applied to the data, this option increases the priority of the selected graph. |
|
|
Descreases the priority of the selected graph. |
|
|
Deletes the selected graph. |
|
|
Sort |
Cycles through the following sorting options for the selected graph: |
|
Displays a window with options for changing the colour for each call value. |
|
|
Sets the zoom level to 1x (normal). There are more zoom options in the following section. |
Genotype Data Points¶
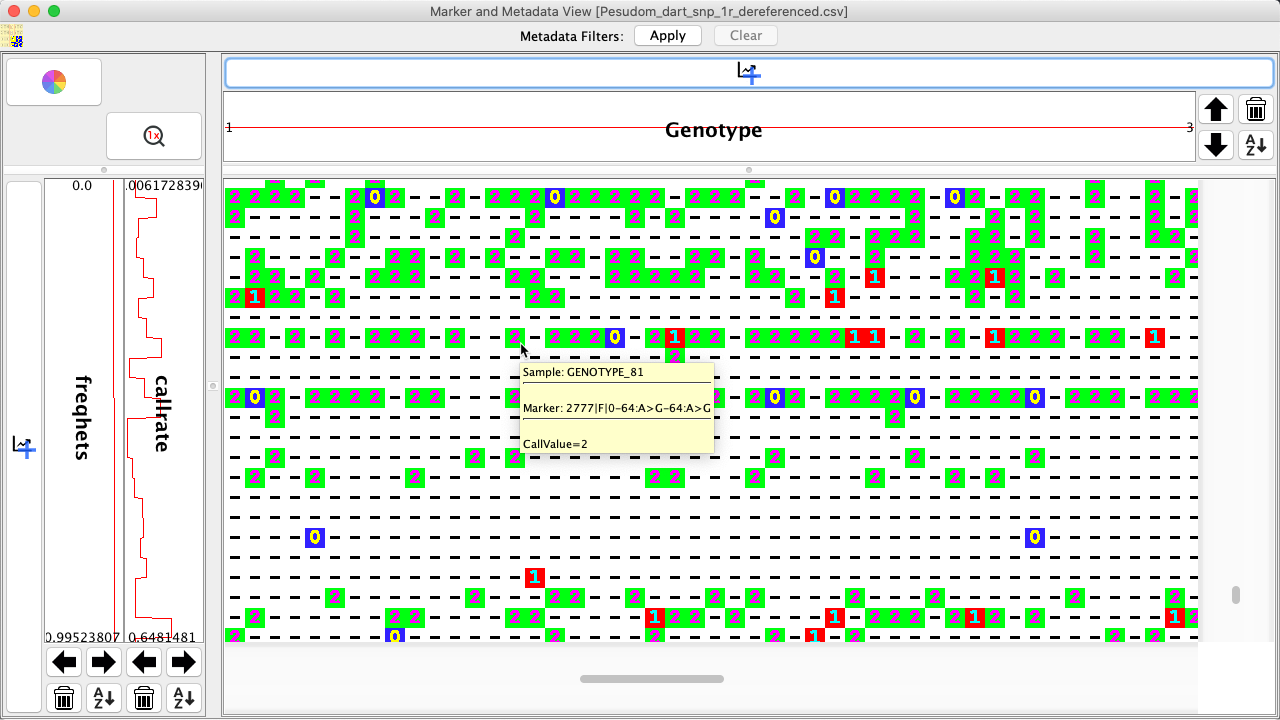
Samples, markers, and call values can be easily viewed in the Marker and Metadata View. Hold the mouse over a genotype data point (this may be easier if zoomed in) to display the sample, marker, and call value as seen in the image below:
Right+clicking on a genotype data point will also provide options which are shown in the image below and listed in the the table below that:
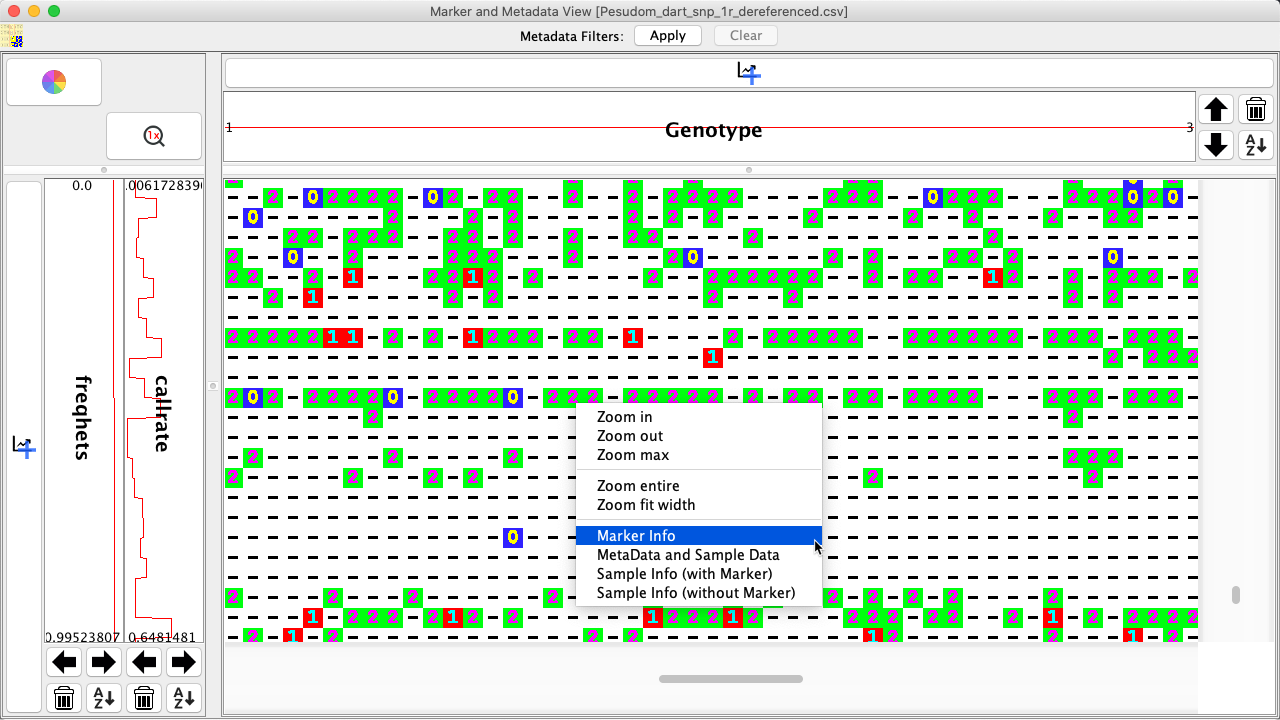
|
Option |
Description |
|---|---|
|
Zoom In |
Zoom in closer to selected call value. |
|
Zoom Out |
Zoom out from selected call value. |
|
Zoom Max |
Maximised the zoom depth (Which will display values as text) onto the focused location |
|
Zoom Entire |
Fits the entire genotypic matrix into view |
|
Zoom Fit Width |
Fits the width of the genotypic matrix into the window Metadata and Sample Data: Shows marker and sample metadata. Need to make it clear which is marker and which is sample |
|
Save as PNG… |
Saves the current Genotypic Data Map with adjacent metadata distributions as a PNG |
|
Show metadata for genotypic data point |
All information for the selected marker which includes anything that was imported to DArTView. |
Exporting Marker Data¶
At any point, you can export your filtered marker data. The Export feature will subset your Marker Data file based on your filters and sorted in the order represented in the Marker and Metadata View.
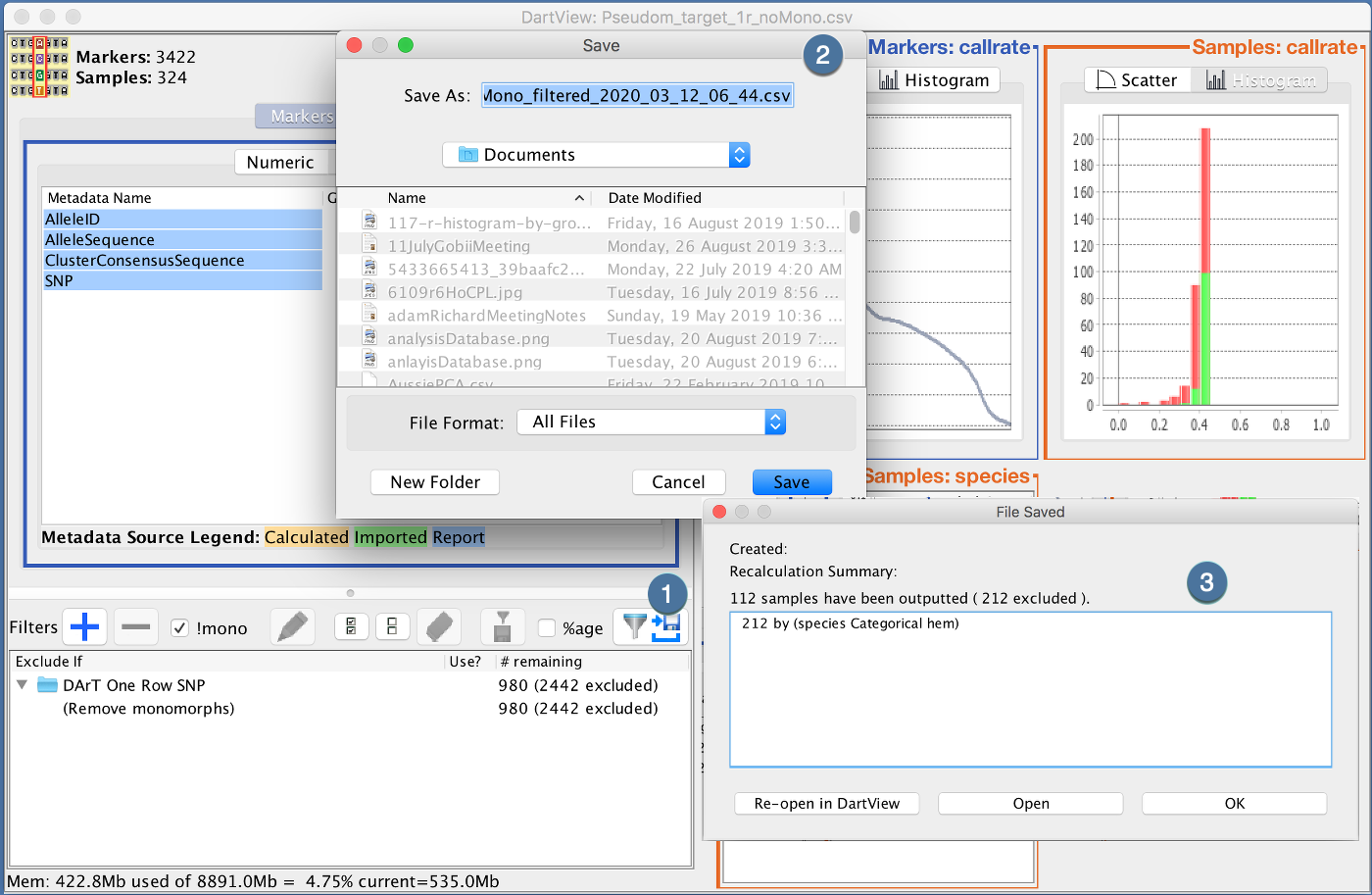
|
Step |
Description |
|---|---|
|
1 |
Click the “Export data applying selected Filters” |
|
2 |
Select a location to save |
|
3 |
Once DArTView has completed exporting your filtered file, you can choose to re-import back in DArTView, open using your default application or continue with the currently loaded marker data file |